The necessary spatiotemporal fine-tuning of podosome structure, turnover and function implies the existence of an intricate network of proteins, comparable to other integrin-based adhesions. However, no systematic effort has yet been made to map the podosome proteome.
Here, we describe the purification of podosome-enriched fractions from primary human macrophages, labeled with isotopically stable amino acids, and the subsequent mass spectrometric analysis of these fractions.
Here, we describe the purification of podosome-enriched fractions from primary human macrophages, labeled with isotopically stable amino acids, and the subsequent mass spectrometric analysis of these fractions.
In order to analyze the podosome proteome and identify novel podosome proteins, we generated podosome-enriched fractions from primary human macrophages, which constitutively form numerous podosomes per cell.
The protocol is based on a two-step method for differential cell lysis, resulting in the removal of the apical cell part, which contains the cytoplasm and nucleus, allowing enrichment of the ventral membrane of cells (“footplates”), containing podosomes.
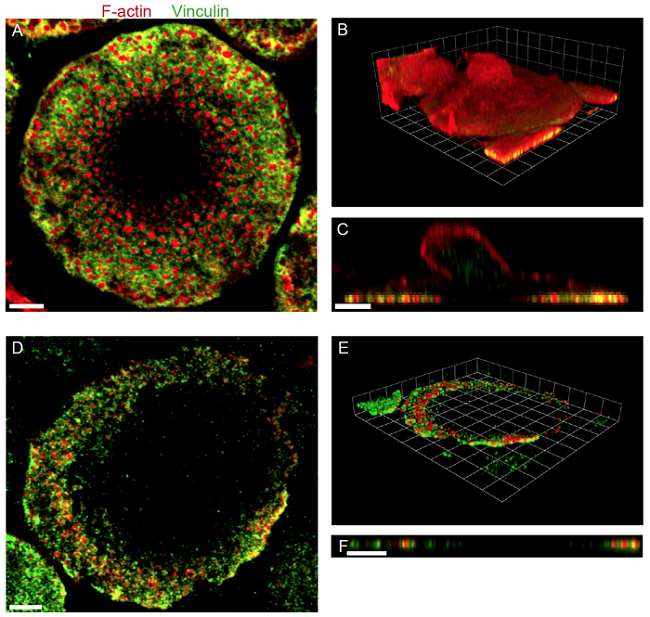
Enrichment of podosomes in the footplate fraction was checked by immunofluorescence labelling of both podosome core and ring components, shown exemplarily for f-actin and vinculin.
The protocol is based on a two-step method for differential cell lysis, resulting in the removal of the apical cell part, which contains the cytoplasm and nucleus, allowing enrichment of the ventral membrane of cells (“footplates”), containing podosomes.
Enrichment of podosomes in the footplate fraction was checked by immunofluorescence labelling of both podosome core and ring components, shown exemplarily for f-actin and vinculin.
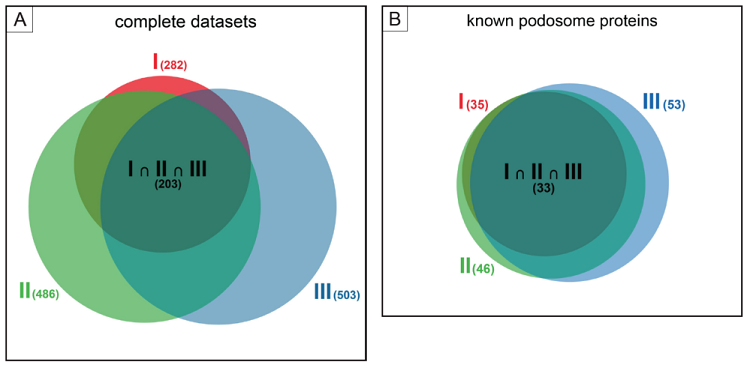
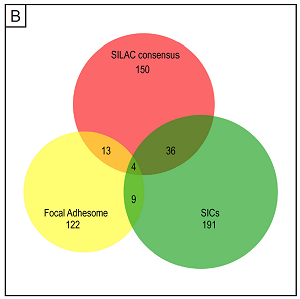
Analysis of podosome-containing versus podosome-free footplates was performed using SILAC (stable isotope labelling of cells) labelling of macrophages, which allows direct comparison between two experimental conditions in a single run of mass spectrometry analysis.
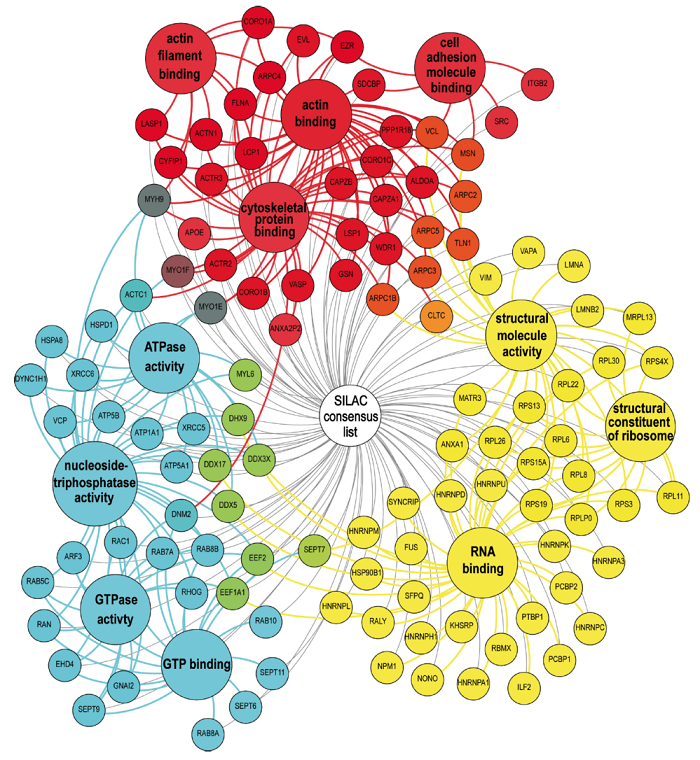
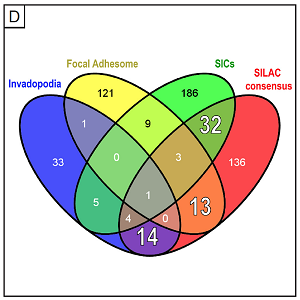
A detailed depiction of the complete network, comprising most of the identified proteins from the consensus list, and grouped according to molecular function. The identified core groups comprise cytoskeletal proteins, especially those binding (f-)actin, cell adhesion molecules, and proteins showing ATPase or GTPase binding,. Two additional (and unexpected) major groups were structural components of ribosomes and proteins showing RNA binding. Localization of the ribonuclear protein hnRNP-K to podosome cores suggests that the presence of these proteins in the podosome fraction is probably not due to contaminations and should be explored further. For the complete list of proteins, use this link.
Individual proteins (small circles) are grouped according to depicted molecular functions (large circles) and color-coded (red, blue, yellow). Proteins that can be attributed to several groups show intermediate colours (orange, purple, green), with final shade (e.g. deep or light orange) depending on the relative attribution to each group. (Cervero et al. EJCB, 2012)